Core functions¶
Core functions.
- permute.core.corr(x, y, alternative='greater', reps=10000, seed=None, plus1=True)[source]¶
Simulate permutation p-value for Pearson correlation coefficient
- Parameters
- xarray-like
- yarray-like
- alternative{‘greater’, ‘less’, ‘two-sided’}
The alternative hypothesis to test
- repsint
- seedRandomState instance or {None, int, RandomState instance}
If None, the pseudorandom number generator is the RandomState instance used by np.random; If int, seed is the seed used by the random number generator; If RandomState instance, seed is the pseudorandom number generator
- plus1bool
flag for whether to add 1 to the numerator and denominator of the p-value based on the empirical permutation distribution. Default is True.
- Returns
- tuple
Returns test statistic, p-value, simulated distribution
- permute.core.one_sample(x, y=None, reps=100000, stat='mean', alternative='greater', keep_dist=False, seed=None, plus1=True)[source]¶
One-sided or two-sided, one-sample permutation test for the mean, with p-value estimated by simulated random sampling with reps replications.
Alternatively, a permutation test for equality of means of two paired samples.
Tests the hypothesis that x is distributed symmetrically symmetric about 0 (or x and y have the same center) against the alternative that x comes from a population with mean
greater than 0 (greater than that of the population from which y comes), if side = ‘greater’
less than 0 (less than that of the population from which y comes), if side = ‘less’
different from 0 (different from that of the population from which y comes), if side = ‘two-sided’
If
keep_dist, return the distribution of values of the test statistic; otherwise, return only the number of permutations for which the value of the test statistic and p-value.- Parameters
- xarray-like
Sample 1
- yarray-like
Sample 2. Must preserve the order of pairs with x. If None, x is taken to be the one sample.
- repsint
number of repetitions
- stat{‘mean’, ‘t’}
The test statistic. The statistic is computed based on either z = x or z = x - y, if y is specified.
If stat == ‘mean’, the test statistic is mean(z).
If stat == ‘t’, the test statistic is the t-statistic– but the p-value is still estimated by the randomization, approximating the permutation distribution.
If stat is a function (a callable object), the test statistic is that function. The function should take a permutation of the data and compute the test function from it. For instance, if the test statistic is the maximum absolute value,
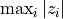 ,
the test statistic could be written:
,
the test statistic could be written:f = lambda u: np.max(abs(u))
- alternative{‘greater’, ‘less’, ‘two-sided’}
The alternative hypothesis to test
- keep_distbool
flag for whether to store and return the array of values of the irr test statistic
- seedRandomState instance or {None, int, RandomState instance}
If None, the pseudorandom number generator is the RandomState instance used by np.random; If int, seed is the seed used by the random number generator; If RandomState instance, seed is the pseudorandom number generator
- plus1bool
flag for whether to add 1 to the numerator and denominator of the p-value based on the empirical permutation distribution. Default is True.
- Returns
- float
the estimated p-value
- float
the test statistic
- list
The distribution of test statistics. These values are only returned if keep_dist == True
- permute.core.spearman_corr(x, y, alternative='greater', reps=10000, seed=None, plus1=True)[source]¶
Simulate permutation p-value for Spearman correlation coefficient
- Parameters
- xarray-like
- yarray-like
- alternative{‘greater’, ‘less’, ‘two-sided’}
The alternative hypothesis to test
- repsint
- seedRandomState instance or {None, int, RandomState instance}
If None, the pseudorandom number generator is the RandomState instance used by np.random; If int, seed is the seed used by the random number generator; If RandomState instance, seed is the pseudorandom number generator
- plus1bool
flag for whether to add 1 to the numerator and denominator of the p-value based on the empirical permutation distribution. Default is True.
- Returns
- tuple
Returns test statistic, p-value, simulated distribution
- permute.core.two_sample(x, y, reps=100000, stat='mean', alternative='greater', keep_dist=False, seed=None, plus1=True)[source]¶
One-sided or two-sided, two-sample permutation test for equality of two means, with p-value estimated by simulated random sampling with reps replications.
Tests the hypothesis that x and y are a random partition of x,y against the alternative that x comes from a population with mean
greater than that of the population from which y comes, if side = ‘greater’
less than that of the population from which y comes, if side = ‘less’
different from that of the population from which y comes, if side = ‘two-sided’
If
keep_dist, return the distribution of values of the test statistic; otherwise, return only the number of permutations for which the value of the test statistic and p-value.- Parameters
- xarray-like
Sample 1
- yarray-like
Sample 2
- repsint
number of repetitions
- stat{‘mean’, ‘t’}
The test statistic.
If stat == ‘mean’, the test statistic is (mean(x) - mean(y)) (equivalently, sum(x), since those are monotonically related)
If stat == ‘t’, the test statistic is the two-sample t-statistic– but the p-value is still estimated by the randomization, approximating the permutation distribution. The t-statistic is computed using scipy.stats.ttest_ind
If stat is a function (a callable object), the test statistic is that function. The function should take two arguments: given a permutation of the pooled data, the first argument is the “new” x and the second argument is the “new” y. For instance, if the test statistic is the Kolmogorov-Smirnov distance between the empirical distributions of the two samples,
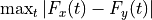 , the test statistic could be written:
, the test statistic could be written:- f = lambda u, v: np.max(
[abs(sum(u<=val)/len(u)-sum(v<=val)/len(v)) for val in np.concatenate([u, v])])
- alternative{‘greater’, ‘less’, ‘two-sided’}
The alternative hypothesis to test
- keep_distbool
flag for whether to store and return the array of values of the irr test statistic
- seedRandomState instance or {None, int, RandomState instance}
If None, the pseudorandom number generator is the RandomState instance used by np.random; If int, seed is the seed used by the random number generator; If RandomState instance, seed is the pseudorandom number generator
- plus1bool
flag for whether to add 1 to the numerator and denominator of the p-value based on the empirical permutation distribution. Default is True.
- Returns
- float
the estimated p-value
- float
the test statistic
- list
The distribution of test statistics. These values are only returned if keep_dist == True
- permute.core.two_sample_conf_int(x, y, cl=0.95, alternative='two-sided', seed=None, reps=10000, stat='mean', shift=None, plus1=True)[source]¶
One-sided or two-sided confidence interval for the parameter determining the treatment effect. The default is the “shift model”, where we are interested in the parameter d such that x is equal in distribution to y + d. In general, if we have some family of invertible functions parameterized by d, we’d like to find d such that x is equal in distribution to f(y, d).
- Parameters
- xarray-like
Sample 1
- yarray-like
Sample 2
- clfloat in (0, 1)
The desired confidence level. Default 0.95.
- alternative{“two-sided”, “lower”, “upper”}
Indicates the alternative hypothesis.
- seedRandomState instance or {None, int, RandomState instance}
If None, the pseudorandom number generator is the RandomState instance used by np.random; If int, seed is the seed used by the random number generator; If RandomState instance, seed is the pseudorandom number generator
- repsint
number of repetitions in two_sample
- stat{‘mean’, ‘t’}
The test statistic.
If stat == ‘mean’, the test statistic is (mean(x) - mean(y)) (equivalently, sum(x), since those are monotonically related)
If stat == ‘t’, the test statistic is the two-sample t-statistic– but the p-value is still estimated by the randomization, approximating the permutation distribution. The t-statistic is computed using scipy.stats.ttest_ind
If stat is a function (a callable object), the test statistic is that function. The function should take two arguments: given a permutation of the pooled data, the first argument is the “new” x and the second argument is the “new” y. For instance, if the test statistic is the Kolmogorov-Smirnov distance between the empirical distributions of the two samples,
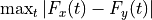 , the test statistic could be written:
, the test statistic could be written:- f = lambda u, v: np.max(
[abs(sum(u<=val)/len(u)-sum(v<=val)/len(v)) for val in np.concatenate([u, v])])
- shiftfloat
The relationship between x and y under the null hypothesis.
If None, the relationship is assumed to be additive (e.g. x = y+d)
A tuple containing the function and its inverse
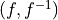 , so
, so
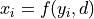 and
and 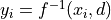
- plus1bool
flag for whether to add 1 to the numerator and denominator of the p-value based on the empirical permutation distribution. Default is True.
- Returns
- tuple
the estimated confidence limits
Notes
- xtolfloat
Tolerance in brentq
- rtolfloat
Tolerance in brentq
- maxiterint
Maximum number of iterations in brentq
- permute.core.two_sample_core(potential_outcomes_all, nx, tst_stat, alternative='greater', reps=100000, keep_dist=False, seed=None, plus1=True)[source]¶
Main workhorse function for two_sample and two_sample_shift
- Parameters
- potential_outcomes_allarray-like
2D array of potential outcomes under treatment (1st column) and control (2nd column). To be passed in from potential_outcomes
- nxint
Size of the treatment group x
- repsint
number of repetitions
- tst_stat: function
The test statistic
- alternative{‘greater’, ‘less’, ‘two-sided’}
The alternative hypothesis to test
- keep_distbool
flag for whether to store and return the array of values of the test statistic. Default is False.
- seedRandomState instance or {None, int, RandomState instance}
If None, the pseudorandom number generator is the RandomState instance used by np.random; If int, seed is the seed used by the random number generator; If RandomState instance, seed is the pseudorandom number generator
- plus1bool
flag for whether to add 1 to the numerator and denominator of the p-value based on the empirical permutation distribution. Default is True.
- Returns
- float
the estimated p-value
- float
the test statistic
- list
The distribution of test statistics. These values are only returned if keep_dist == True
- permute.core.two_sample_shift(x, y, reps=100000, stat='mean', alternative='greater', keep_dist=False, seed=None, shift=None, plus1=True)[source]¶
One-sided or two-sided, two-sample permutation test for equality of two means, with p-value estimated by simulated random sampling with reps replications.
Tests the hypothesis that x and y are a random partition of x,y against the alternative that x comes from a population with mean
greater than that of the population from which y comes, if side = ‘greater’
less than that of the population from which y comes, if side = ‘less’
different from that of the population from which y comes, if side = ‘two-sided’
If
keep_dist, return the distribution of values of the test statistic; otherwise, return only the number of permutations for which the value of the test statistic and p-value.- Parameters
- xarray-like
Sample 1
- yarray-like
Sample 2
- repsint
number of repetitions
- stat{‘mean’, ‘t’}
The test statistic.
If stat == ‘mean’, the test statistic is (mean(x) - mean(y)) (equivalently, sum(x), since those are monotonically related)
If stat == ‘t’, the test statistic is the two-sample t-statistic– but the p-value is still estimated by the randomization, approximating the permutation distribution. The t-statistic is computed using scipy.stats.ttest_ind
If stat is a function (a callable object), the test statistic is that function. The function should take two arguments: given a permutation of the pooled data, the first argument is the “new” x and the second argument is the “new” y. For instance, if the test statistic is the Kolmogorov-Smirnov distance between the empirical distributions of the two samples,
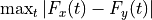 , the test statistic could be written:
, the test statistic could be written:- f = lambda u, v: np.max(
[abs(sum(u<=val)/len(u)-sum(v<=val)/len(v)) for val in np.concatenate([u, v])])
- alternative{‘greater’, ‘less’, ‘two-sided’}
The alternative hypothesis to test
- keep_distbool
flag for whether to store and return the array of values of the irr test statistic
- seedRandomState instance or {None, int, RandomState instance}
If None, the pseudorandom number generator is the RandomState instance used by np.random; If int, seed is the seed used by the random number generator; If RandomState instance, seed is the pseudorandom number generator
- shiftfloat
The relationship between x and y under the null hypothesis.
A constant scalar shift in the distribution of y. That is, x is equal in distribution to y + shift.
A tuple containing the function and its inverse
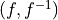 , so
, so
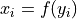 and
and 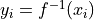
- plus1bool
flag for whether to add 1 to the numerator and denominator of the p-value based on the empirical permutation distribution. Default is True.
- Returns
- float
the estimated p-value
- float
the test statistic
- list
The distribution of test statistics. These values are only returned if keep_dist == True